Web Servers
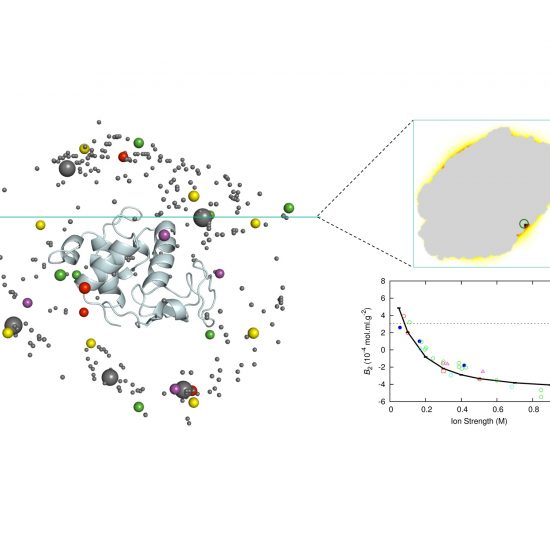
FMAPB2
FMAPB2 implements FMAP, a method based on fast Fourier transform (FFT), to calculate second virial coefficients (B2) for proteins represented at the all-atom level in implicit solvent.
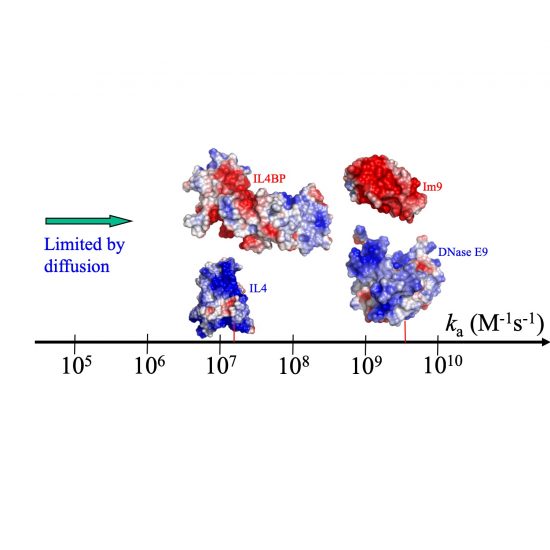
TransComp
TransComp implements the Transient-Complex theory for predicting protein-protein and protein-RNA association rate constants (ka). This predictor provides critical missing kinetic information for quantitative modeling of protein interaction networks.
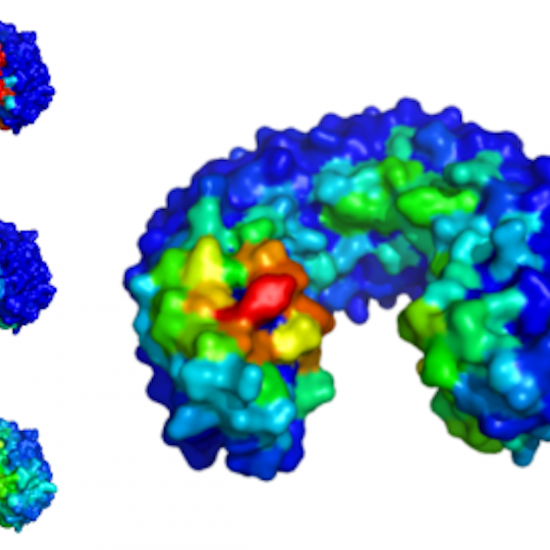
meta-PPISP
meta-PPISP is built on three individual web servers: cons-PPISP, PINUP, and Promate. Cross validation showed that meta-PPISP outperforms all the three individual servers. At coverages identical to those of the individual methods, the accuracy of meta-PPISP is higher by 4.8 to 18.2 percentage points.
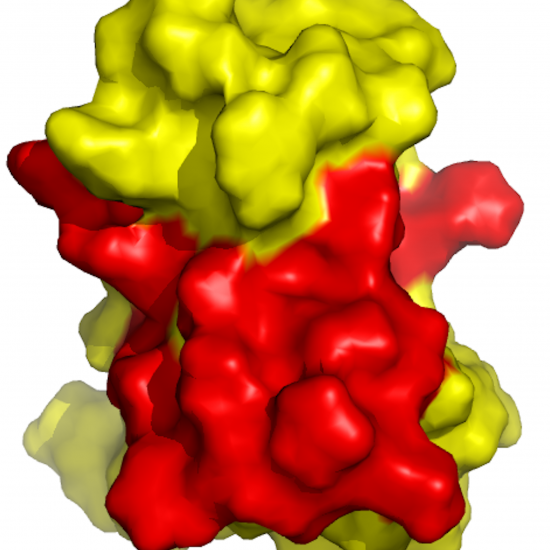
cons-PPISP
cons-PPISP is a consensus neural-network Protein-Protein Interaction Site Predictor. The input is the unbound structure of a protein, which is known to bind another protein. The prediction can be used to drive docking of the protein-protein complex or to assist the scoring of docked structures.
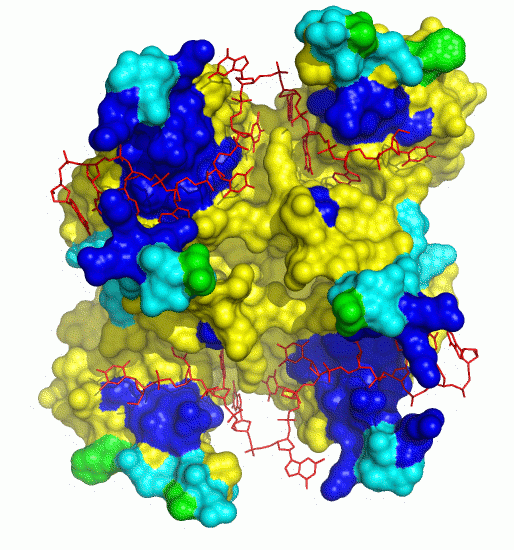
DISPLAR
DISPLAR is a DNA-Interaction Site Predictor, with data such as sequence profiles of a List of Adjacent Residues as input. The predictions of cons-PPISP and DISPLAR can be used to build structural models for multi-component protein-DNA complexes.
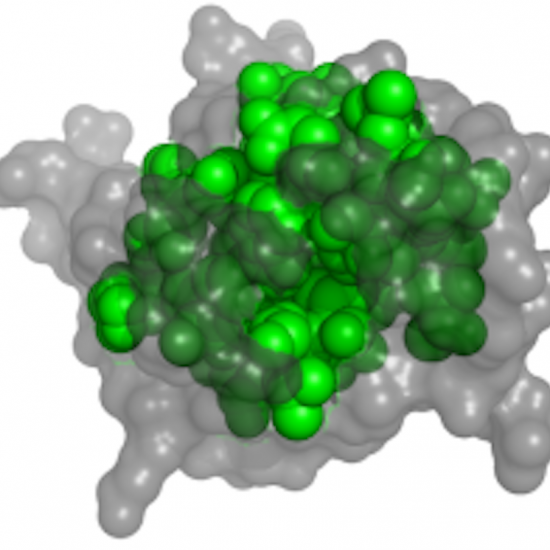
WESA
WESA is a meta-predictor, based on a Weighted Ensemble of five methods, for Solvent Accessibility of residues, using the protein sequence as input. It has an expected accuracy of 80%. The prediction can be used for structure prediction.